Note
Go to the end to download the full example code.
Barnes Filter#
Barnes filter is a commonly used spatial filtering method that mainly uses two constants g and c to calculate Gaussian weights, and performs spatial interpolation for each grid point, thus becoming a low-pass filter that filters out high-frequency fluctuations. When using two different schemes of constant g and c schemes, both retain low-frequency fluctuations of different scales. The difference between the filtering results of the two methods can result in mesoscale fluctuations.
See also
Maddox, R. A. (1980). An Objective Technique for Separating Macroscale and Mesoscale Features in Meteorological Data. Monthly Weather Review, 108(8), 1108-1121. https://journals.ametsoc.org/view/journals/mwre/108/8/1520-0493_1980_108_1108_aotfsm_2_0_co_2.xml
Before proceeding with all the steps, first import some necessary libraries and packages
import numpy as np
import cartopy.crs as ccrs
import easyclimate as ecl
Open tuturial dataset
hgt_data = ecl.open_tutorial_dataset("hgt_2022_day5").hgt.sel(level = 1000)
hgt_data
hgt_2022_day5.nc ━━━━━━━━━━━━━━━━━━━━━ 100.0% • 1.7/1.7 MB • 18.1 MB/s • 0:00:00
Filter dataset using easyclimate.filter.calc_barnes_lowpass
Caculating the first revision...
Caculating the second revision...
Draw results and differences.
fig, ax = ecl.plot.quick_draw_spatial_basemap(3, 1, figsize=(5, 12), central_longitude=180)
axi = ax[0]
hgt_data.isel(time = 0).plot.contourf(
ax = axi, levels = 21,
transform = ccrs.PlateCarree(),
cbar_kwargs={"location": "bottom", "aspect": 30, "label": ""},
)
axi.set_title("Raw data")
axi = ax[1]
hgt_data1.isel(time = 0).plot.contourf(
ax = axi, levels = 21,
transform = ccrs.PlateCarree(),
cbar_kwargs={"location": "bottom", "aspect": 30, "label": ""},
)
axi.set_title("Filtered data")
axi = ax[2]
draw_dta = hgt_data.isel(time = 0) - hgt_data1.isel(time = 0)
draw_dta.plot.contourf(
ax = axi, levels = np.linspace(-30, 30, 21),
transform = ccrs.PlateCarree(),
cbar_kwargs={"location": "bottom", "aspect": 30, "label": ""},
)
axi.set_title("Difference: Mesoscale fluctuations")
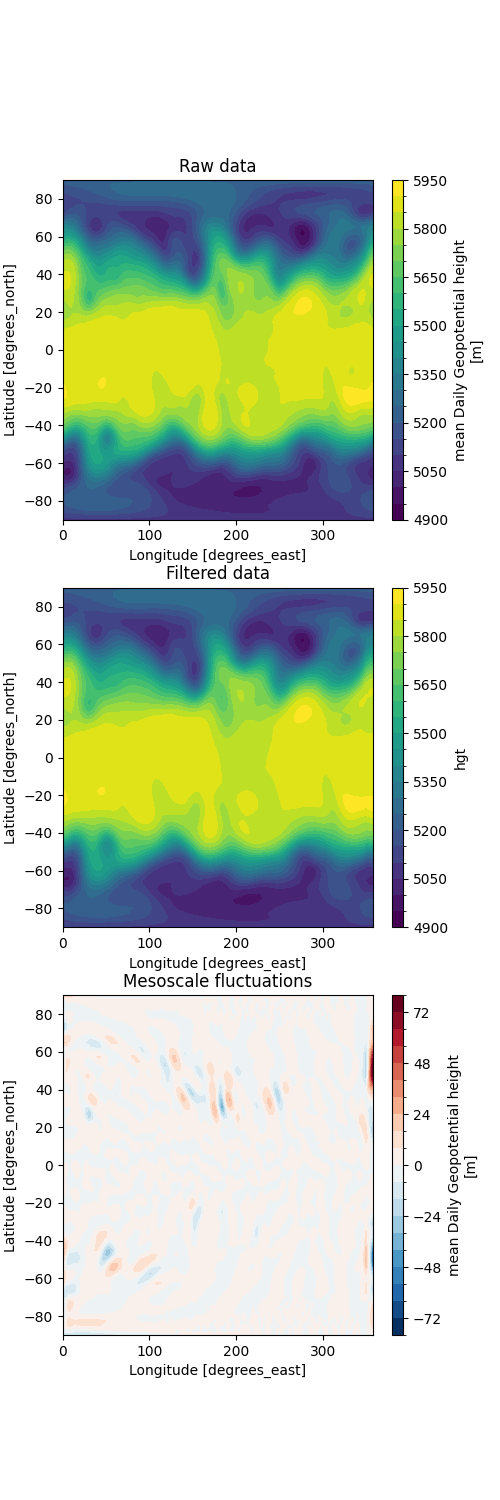
Text(0.5, 1.0, 'Difference: Mesoscale fluctuations')
Total running time of the script: (0 minutes 5.271 seconds)